Cell proliferation is the basis of growth, development, reproduction and inheritance of organisms. It is one of the important physiological functions of living cells and an important life characteristic of organisms. Cell proliferation is an important indicator for evaluating cell metabolism, physiological and pathological conditions. Common detection methods include BrdU staining, EdU staining and clone formation.
I. BrdU/EdU staining
1. Experimental principle
BrdU (5-Bromo-2'-deoxuridine) is a derivative of thymine, which can replace thymine and selectively integrate into newly synthesized DNA (S phase of cell cycle). This incorporation can exist stably and enter daughter cells with DNA replication. BrdU-specific antibodies can be used to detect the incorporated BrdU and then judge the proliferation ability of cells. EdU (5-Ethynyl-2'-deoxyuridine) is a thymine nucleoside analog that can replace thymine and be incorporated into the synthesized DNA molecules during DNA replication. Based on the specific reaction of Apollo® fluorescent dye and EdU, the proliferation ability of cells can be directly detected. Although both BrdU and Edu methods can detect cell proliferation, they are different. BrdU antibodies are larger, and DNA must be partially denatured to form single strands before they can bind to BrdU and complete the detection; EdU is only 1/500 of the size of BrdU antibodies, and is easier to diffuse in cells. It can be effectively detected without DNA denaturation, which can effectively avoid sample damage, and has higher sensitivity and faster detection speed.
2. Experimental principle diagram
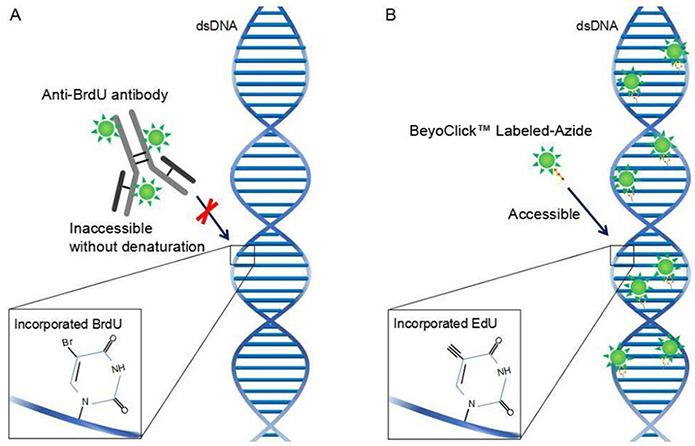
3. Experimental steps
3.1 BrdU staining
3.1.1 Take cells in the logarithmic growth phase and inoculate them into a 96-well plate at 1×105 cells per well (the specific density depends on the cell growth rate). After the cells return to normal, perform the required drug treatment or other stimulation treatment;
3.1.2 Add the prepared 10× BrdU solution to the plate wells to obtain a final concentration of 1×.
3.1.3 Place the cells in a thermostat. The general incubation time is 1-24 h;
3.1.4 Remove the culture medium. If the cells are suspended, centrifuge at 300 g for 10 min, then discard the culture medium;
3.1.5 Add Fixing/Denaturing Solution at 100 μL/well, let the plate stand at room temperature for 30 min, and discard the solution;
3.1.6 Add 1×BrdU antibody solution at 100 μL/well, let the plate stand at room temperature for 1 h;
3.1.7 Discard the solution and wash 3 times with 1× Wash Buffer;
3.1.8 Add 1×HRP-labeled secondary antibody solution at 100 μL/well, let the plate stand at room temperature for 30 min, and discard the solution; wash the plate 3 times with 1× Wash Buffer.
3.1.9 Add 100 μL TMB Substrate;
3.1.10 Incubate at room temperature for 30 min;
3.1.11 Add 100 μL STOP Solution;
3.1.12 Read the absorbance at 450 nm (for the best reading, read the plate within 30 min of adding STOP Solution).
3.2 EdU staining
3.2.1 EdU labeling, fixation, washing and permeabilization of cultured cells
3.2.1.1 Take cells in logarithmic growth phase and inoculate them into 96-well plates at 1×105 cells per well (the specific density depends on the growth rate of the cells). After the cells return to normal, perform the required drug treatment or other stimulation treatment;
3.2.1.2 Prepare 2× EdU working solution, preheat at 37℃, and add equal volume to the 96-well plate to make the final EdU concentration in the 96-well plate 1×. Replacing all the culture medium may affect cell proliferation, so it is not recommended to replace all the culture medium;
3.2.1.3 Continue to incubate the cells for 2 hours. The length of the incubation time depends on the cell growth rate, and the incubation time is usually about 10% of the cell cycle;
3.2.1.4 After EdU labeling of cells, discard the culture medium and add 1 mL of fixative solution to fix at room temperature for 15 minutes.
3.2.1.5 Discard the fixative solution and wash the cells 3 times with 1 mL of washing solution per well, 5 minutes each time.
3.2.1.6 Discard the washing solution and incubate each well with 1 mL of permeabilization solution at room temperature for 15 minutes.
3.2.1.7 Discard the permeabilization solution and wash the cells 2 times with 1 mL of washing solution per well, 5 minutes each time.
3.2.2 EdU detection
The reaction system in each well of the 96-well plate is 50 μL of reaction mixture, and the corresponding reaction mixture systems for 6-, 12-, 24-, 48- and 384-well plates are 500 μL, 200 μL, 100 μL, 70 μL and 20 μL, respectively.
3.2.2.1 Remove the washing solution in the above wells, add 50 μL of Click reaction solution to each well (refer to the component sequence and volume in the table below to prepare the Click reaction solution, and use it within 15 minutes after preparation), and gently shake the culture plate to ensure that the reaction mixture can evenly cover the sample;
3.2.2.2 Incubate at room temperature in the dark for 30 minutes.
3.2.2.3 Aspirate the Click reaction solution and wash 3 times with washing solution, 5 minutes each time.
3.3.3 Nuclear staining
In order to detect the proportion of cell proliferation, you can consider using Hoechst 33342 for nuclear staining.
3.3.3.1 After removing the washing solution, add 1 mL of 1× Hoechst 33342 solution to each well and incubate at room temperature in the dark for 10 minutes.
3.3.3.2 Remove the 1× Hoechst 33342 solution.
3.3.3.3 Wash with washing solution 3 times, 5 minutes each time.
3.3.3.4 Then perform fluorescence detection. Hoechst 33342 is a blue fluorescent light with a maximum excitation wavelength of 346 nm and a maximum emission wavelength of 460 nm.
4. Result Example

EdU marks cells undergoing DNA replication, which appear green under a fluorescent microscope; Hoechst 33342 marks the nucleus of cells, which appear blue under a microscope. The green marker is NeuN.
II. Cell clone formation experiment
1. Experimental principle
Cell clone formation experiment is an important technical method used to detect cell proliferation ability, invasiveness, sensitivity to killing factors and other projects. The so-called clone is a cell group composed of the offspring of a single cell that proliferates for more than 6 generations in vitro. The cell clone formation rate indicates the number of cells that survive and form clones after inoculation, reflecting the dependence and proliferation ability of the cell group. Due to different cell biological characteristics, the clone formation rate varies greatly. Generally, normal cells have a weak clone formation rate, while tumor cells have a strong clone formation rate. Clone formation is divided into two categories: plate clone and soft agar clone according to the different culture media used. Plate clone formation experiment is mainly used for cells that adhere to the wall. Soft agar clone formation experiment is mainly used for suspended tumor cells and transformed cell lines.
2. Experimental principle diagram

3. Experimental steps
3.1 Plate clone formation
3.1.1 After digesting the cells in the logarithmic growth phase with trypsin, resuspend them in complete medium (basal medium + 10% fetal bovine serum) into a cell suspension and count them;
3.1.2 Inoculate 400-1000 cells/well in the culture plate (6-well plate) (determined according to the cell growth situation, generally 700 cells/well);
3.1.3 Continue to culture until 14 days or until the number of cells in most single clones is greater than 50, change the medium every 3 days and observe the cell status;
3.1.4 Discard the culture medium, wash once with PBS, add 1 mL 4% paraformaldehyde to each well and fix for 30-60 min;
3.1.5 Wash once with PBS, add 1 mL crystal violet stain to each well, and stain the cells for 10-20 min;
3.1.6 Wash the cells several times with PBS, dry them, and take pictures with a camera (take pictures of the entire six-well plate and each well separately);
3.2 Soft agar clone formation
3.2.1 Prepare 1.2% and 0.7% agarose, sterilize them by high pressure, and keep them at 42 ℃ to keep them melted;
3.2.2 Mix 1.2% agarose gel with 2× culture medium 1:1, spread it on a 6-well plate as the lower layer of gel, 1.5 mL per well, and wait for it to solidify at room temperature;
3.2.3 After trypsin digestion of cells in the logarithmic growth phase, resuspend them in complete culture medium (basal culture medium + 10% fetal bovine serum) into a cell suspension, and adjust the cell concentration to 5×104/mL;
3.2.4 Mix 0.7% agarose gel with 2× culture medium 1:1, add 100 μL of cell suspension, mix well, and add it to a 6-well plate as the upper layer of gel, 1.5 mL per well.
3.2.5 After solidification, add culture medium on top and replace it every 3 days;
3.2.6 Observe the size of clones under a microscope after culturing for 2-3 weeks according to the growth rate of cells. As with plate clones, take photos under a microscope if each clone is >50 cells. If the entire well is photographed, add 200 μL of nitro blue tetrazolium chloride (NBT) to each well for staining, incubate at 37°C overnight, and take photos.
4. Example of experimental results
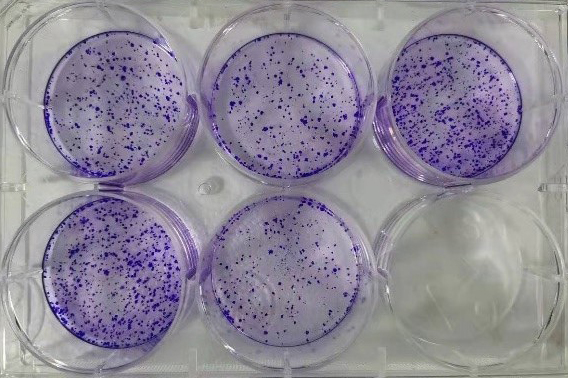
Plate clone formation. After the cell clones are formed, they turn blue after being stained with crystal violet; under the same number of inoculated cells, the more cell clones there are, the stronger the cell proliferation ability is.
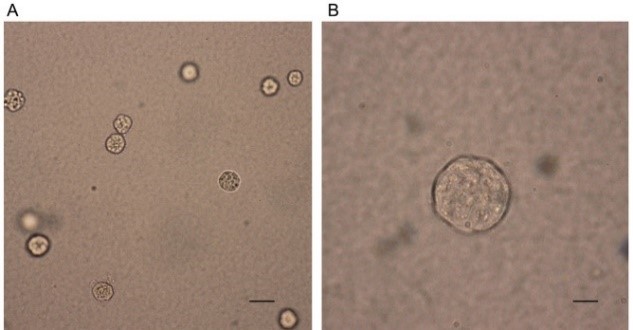
Soft agar colony formation. Dispersed cell clones can be seen under a microscope, and the ratio of the number of clones to the number of inoculated cells can measure the cell proliferation ability; the higher the ratio, the stronger the proliferation ability.
References
[1] Matatall KA, Kadmon CS, King KY. Detecting Hematopoietic Stem Cell Proliferation Using BrdU Incorporation. Methods Mol Biol. 2018;1686:91-103.
[2] Crane AM, Bhattacharya SK. The use of bromodeoxyuridine incorporation assays to assess corneal stem cell proliferation. Methods Mol Biol. 2013;1014:65-70.
[3] Diermeier-Daucher S, Clarke ST, Hill D, Vollmann-Zwerenz A, Bradford JA, Brockhoff G. Cell type specific applicability of 5-ethynyl-2'-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Cytometry A. 2009 Jun;75(6):535-46.
[4] Angelozzi M, de Charleroy CR, Lefebvre V. EdU-Based Assay of Cell Proliferation and Stem Cell Quiescence in Skeletal Tissue Sections. Methods Mol Biol. 2021;2230:357-365.
[5] Tang H, Liu J, Huang J. GMFG (glia maturation factor gamma) inhibits lung cancer growth by activating p53 signaling pathway. Bioengineered. 2022 Apr;13(4):9284-9293.
[6] Du F, Zhao X, Fan D. Soft Agar Colony Formation Assay as a Hallmark of Carcinogenesis. Bio Protoc. 2017 Jun 20;7(12):e2351.