The cell cycle refers to the process from the formation of daughter cells in one cell division to the formation of daughter cells in the next cell division. The formation of daughter cells is usually regarded as a sign of the end of a cell division. The life of a cell begins with the division of the mother cell that produces it, and ends with the formation of its daughter cells, or the death of the cell itself. Apoptosis, or programmed cell death, is one of the built-in defense mechanisms of cells. It plays an important role in the normal development and disease resistance of organisms. It involves the activation, expression and regulation of a series of genes. It is a death process that cells actively strive for in order to better adapt to the environment. It is specifically manifested as: budding to form apoptotic bodies, cutting of DNA between nucleosomes, phagocytosis and digestion of apoptotic bodies, and other continuous processes.
I. Cell cycle
1. Experimental principle
Flow cytometry uses monoclonal antibodies labeled with different fluorescent substances to interact with the components to be tested, and then detects the cells to be tested on a flow cytometer. The cells to be tested are arranged in a single row with the flowing sheath fluid in the flow chamber, and quickly pass through the laser focusing area one by one. When the laser irradiates each cell, two scattered lights, forward angle scattering and side angle scattering, as well as the signal emitted by the excited fluorescent marker, can be obtained at the same time. Using these signals, the relative content can be calculated, thereby obtaining the relative ratio of the cell population. The cell cycle is divided into two stages: interphase and division phase. The interphase is further divided into three phases: the pre-DNA synthesis phase (G1 phase), the DNA synthesis phase (S phase) and the late DNA synthesis phase (G2 phase). Some cells temporarily leave the cell cycle after the division, stop cell division, and perform certain biological functions (G0 phase). Due to the different DNA content in each phase of the cell cycle, the G1/G0 phase of normal cells usually has the DNA content of diploid cells (2N), while the G2/M phase has the DNA content of tetraploid cells (4N), and the DNA content of the S phase is between diploid and tetraploid. PI can bind to DNA, and its fluorescence intensity directly reflects the DNA content in the cell. Therefore, when the intracellular DNA content is detected by flow cytometry PI staining, the phases of the cell cycle can be divided into G1/G0 phase, S phase and G2/M phase.
2. Cell cycle experimental steps
2.1 Cell plating treatment
2.1.1 First, 14 porous materials were placed in the basic culture medium and soaked for 60 min.
2.1.2 Digested with 0.25% trypsin at 37 ℃, counted the cells, and adjusted the cell concentration to 5×104/μL.
2.1.3 After soaking, placed in a 24-well ultra-low attachment culture plate, and 100 μL of cell suspension was added to each scaffold and allowed to stand for 30 min.
2.1.4 Use a pipette to aspirate the remaining culture solution in the culture well and add it again on the porous scaffold. Repeat 4 times so that the cells can be fully and evenly adhered to the porous scaffold.
2.1.5 After inoculation, place the culture plate in the incubator for pre-incubation for 2 h, waiting for the initial adhesion of cells.
2.1.6 Add basal culture medium to each well again to cover the material and place in the incubator for incubation.
2.1.7 After the incubation time is up, digest with trypsin, centrifuge at 1000 g for 5 min to collect the cells. Add 1 mL of pre-cooled PBS to resuspend, centrifuge the cells again, and carefully remove the supernatant.
2.1.8 Add 1 mL of pre-cooled 70% ethanol to resuspend the cells, and fix them at 4 ℃ for 24 h.
2.1.9 Centrifuge at 1000 g for 5 min to collect the cells, remove the supernatant, add 1 mL of pre-cooled PBS to resuspend, centrifuge again, and remove the supernatant.
2.1.10 Prepare propidium iodide staining solution according to the table below:
| Reagents | Each sample |
| Staining Buffer | 0.5 mL |
| Propidium iodide staining solution(20×) | 25 μL |
| RNase A (50×) | 10 μL |
2.1.11 Staining: Add 0.5 mL of propidium iodide staining solution to each tube of cell sample, slowly and fully resuspend the cell pellet, and incubate at 37 oC in the dark for 30 min.
2.1.12 Flow cytometry: Use a flow cytometer to detect red fluorescence at an excitation wavelength of 488 nm and detect light scattering at the same time.
2.1.13 Use CellQuest software to analyze the cell cycle.
3. Example of experimental results
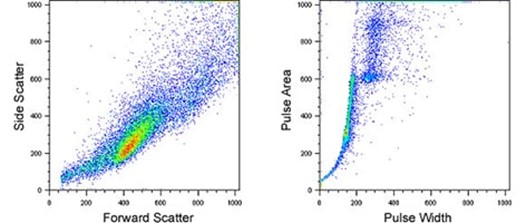
Forward scatter and side scatter data plots for identifying individual cells; pulse shape analysis plots for identifying cell clusters and adherent cells;
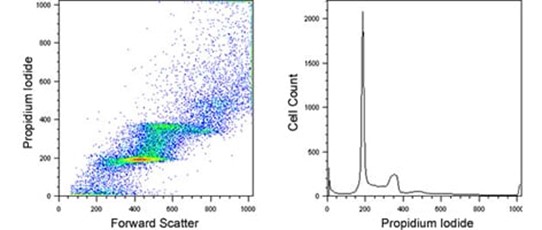
Bottom right: Forward scatter - PI signal plot; PI histogram.
II. Cell apoptosis
1. Experimental principle
Annexin V and PI double staining is a classic method for flow cytometry detection of cell apoptosis. It is based on the principle that phosphatidylserine (PS) on the cell membrane in the early stage of apoptosis flips from the inner side of the cell membrane to the surface of the cell membrane. Annexin V is a Ca2+-dependent phospholipid binding protein with a molecular weight of 35-36 kDa, which can bind to PS with high affinity. Annexin V is labeled with fluorescein FITC, and the occurrence of cell apoptosis can be detected using a flow cytometer or a fluorescence microscope. Propidium iodide (PI) is a dye that can bind to DNA. It cannot penetrate the intact cell membrane of normal cells or early apoptotic cells, but in cells in the middle and late stages of apoptosis and dead cells, PI can penetrate the cell membrane and dye the cell nucleus red. Therefore, when Annexin V is used in combination with PI, it can be used to identify living cells, apoptotic cells and dead cells.
2. Cell apoptosis experimental steps
2.2.1 After digesting the cells with EDTA-free trypsin, centrifuge at 300 g and 4 ℃ for 5 min to collect the cells.
2.2.2 Wash the cells twice with pre-cooled PBS, centrifuge at 300 g and 4 ℃ for 5 min each time.
2.2.3 Aspirate the PBS and add 100 μL 1×Bingding Buffer to resuspend the cells.
2.2.4 Add 5 μL Annexin-V-FITC and 10 μL PI Staining Solution for double staining.
2.2.5 Protect from light and react at room temperature for 15 min.
2.2.6 Add 400 μL 1×Bingding Buffer, mix well, place on ice and immediately detect with flow cytometer.
2.2.7 Use CellQuest software to analyze the proportion of cell apoptosis.
3. Example of experimental results
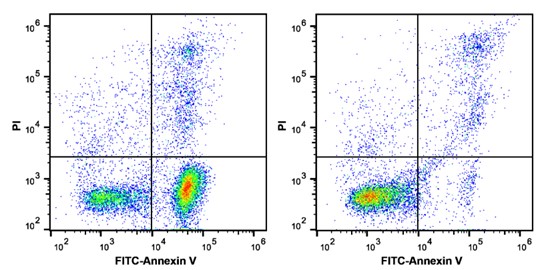
The results of flow cytometry detection of the experimental group (left) and the control group (right) by Annexin V and PI double staining: Annexin V-FITC single positive cells are early apoptotic cells, Annexin V-FITC and PI double positive cells are necrotic or late apoptotic cells, and PI single positive cells are naked nucleus cells.
References
[1] Rieger A M, Nelson K L, Konowalchuk J D, et al. Modified Annexin V/Propidium Iodide Apoptosis Assay For Accurate Assessment of Cell Death[J]. Journal of Visualized Experiments Jove, 2011(50).
[2] Crowley L C, Marfell B J, Scott A P, et al. Quantitation of Apoptosis and Necrosis by Annexin V Binding, Propidium Iodide Uptake, and Flow Cytometry[J]. Cold Spring Harbor Protocols, 2016, 2016(11):pdb.prot087288.
[3] Rieger A M, Barreda D R. Accurate Assessment of Cell Death by Imaging Flow Cytometry[M]. Methods Mol Biol, 2016.
[4] Sawai H, Domae N. Discrimination between primary necrosis and apoptosis by necrostatin-1 in Annexin V-positive/propidium iodide-negative cells[J]. Biochemical & Biophysical Research Communications, 2011, 411(3):569-573.
[5] Pietkiewicz S, Schmidt J H, Lavrik I N. Quantification of apoptosis and necroptosis at the single cell level by a combination of Imaging Flow Cytometry with classical Annexin V/propidium iodide staining[J]. Journal of Immunological Methods, 2015, 423:99-103.