qPCR (Quantitative Real-time PCR) is a real-time fluorescence quantitative PCR technology that uses intercalating dyes or fluorescent probes to monitor the fluorescence intensity during the PCR process to compare the DNA levels between samples, and finally uses a standard curve to perform quantitative analysis on unknown templates. According to the principle of chemiluminescence, it can be divided into: fluorescent dye method and TaqMan probe method.
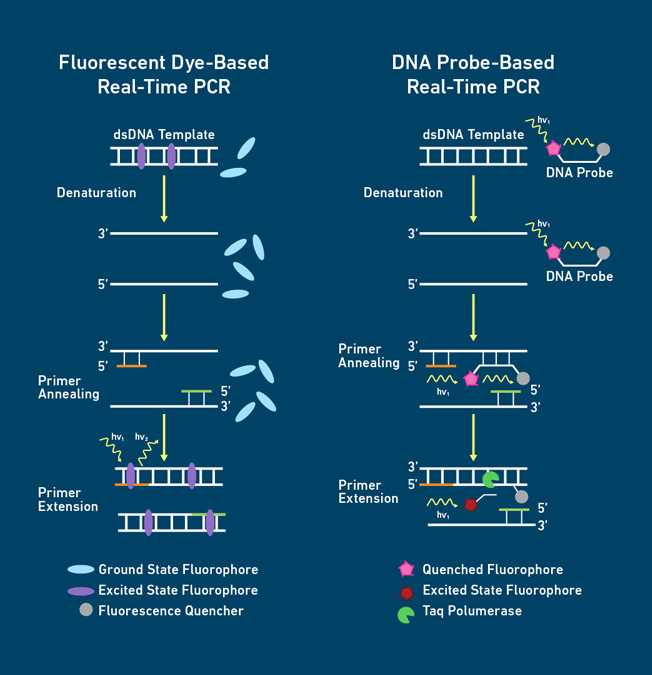
1. Experimental principle
1.1 Fluorescent dye method
In the PCR reaction system, an excess of fluorescent dye is added. After the fluorescent dye is specifically incorporated into the DNA double strand, it emits a fluorescent signal, while the dye molecules that are not incorporated into the strand will not emit any fluorescent signal, thereby ensuring that the increase of the fluorescent signal is completely synchronized with the increase of the PCR product.
1.2 TaqMan probe method
During PCR amplification, a pair of primers are added and a specific TaqMan fluorescent probe is also added. The probe specifically binds to the template, and its binding site is between the two primers. When the probe is complete, the spatial distance between the reporter fluorescent group and the quencher fluorescent group is very close, and the fluorescent signal emitted by the reporter fluorescent group will be absorbed by the quencher fluorescent group, so that the instrument cannot detect the fluorescent signal. During the PCR extension phase, Taq DNA polymerase synthesizes new chains along the template chain from 5' to 3'. When Taq DNA polymerase reaches the probe binding site, the 5'-3' exonuclease activity of Taq DNA polymerase cuts off the reporter fluorescent group connected to the 5' end of the probe, separating the reporter fluorescent group and the quencher fluorescent group, thereby emitting fluorescence. The number of fluorescent molecules cut is proportional to the number of PCR products. Therefore, the purpose of detecting the amplification amount of PCR products can be achieved by detecting the fluorescence intensity in the PCR reaction system.
2. Experimental steps
2.1 Total RNA extraction
2.1.1 Cell lysis or tissue homogenization
Adherent cells: Aspirate the culture medium, generally add 1 mL Trizol to each well of a six-well plate, and add 0.5 mL Trizol to each well of a 12-well plate. Shake 3-5 times, then pipette several times to ensure complete lysis, and then transfer to a 1.5 mL centrifuge tube.
Suspended cells: Collect cells by centrifugation, aspirate the liquid, and add 1 mL Trizol for every 5×106 cells. Use a gun to blow and beat to ensure complete lysis.
Tissue: Put the tissue block in a mortar, add a small amount of liquid nitrogen, grind quickly, and when the tissue becomes soft, add a small amount of liquid nitrogen and grind again. Repeat this three times. Add 1 mL Trizol for every 50-80 mg of tissue and transfer it to a centrifuge tube. Place the lysis solution with Trizol at room temperature for 5 minutes, centrifuge at 4 ℃ 12,000 rpm for 10 minutes, and pipette the clarified Trizol lysis product (supernatant) into a new centrifuge tube to remove insoluble matter or greasy floating matter.
2.1.2 Add 0.2 mL chloroform for every 1 mL Trizol used, shake vigorously for 15 seconds, place at room temperature for 3 minutes, and centrifuge at 4 ℃ 12,000 rpm for 15 minutes.
2.1.3 Transfer the upper aqueous phase to a new tube and precipitate the RNA in the aqueous phase with isopropanol. Add 0.5 mL isopropanol for every 1 mL Trizol used, invert several times to mix, and place in a -20 ℃ refrigerator for 30 minutes. If extracting small RNA such as microRNA, place it at -80 ℃ for precipitation overnight.
2.1.4 Centrifuge at 4 ℃ 12 000 rpm for 30 min, and RNA precipitate can be seen at the bottom of the tube.
2.1.5 Discard the supernatant and wash the RNA precipitate with 75% ethanol (prepared with DEPC water). Add at least 1 mL of 75% ethanol for every 1 mL of Trizo1 used. Centrifuge at 4 ℃, 7500 rpm for 5 min.
2.1.6 Place the dry RMA precipitate at room temperature for about 3-5 min. Add an appropriate amount of RNase-free water to dissolve the RNA and then test the RNA concentration and purity.
2.2 cDNA synthesis
Reverse transcription system
Reagents | Usage (μL) |
RNA | ≤ 1 μg |
Enzyme Mix | 1 |
5×All-in-one qRT SuperMix | 4 |
RNase Free dH2O | Up to 20 |
The above reaction system was reacted in a PCR instrument at 50°C for 15 min for reverse transcription, and then reacted at 85°C for 5 s to inactivate the RT enzyme; the obtained cDNA product could be stored at -80°C for future use.
2.3 Real Time PCR
2.3.1 Primer design is done at NCBI and must be synthesized before the experiment.
2.3.2 Quantitative PCR reaction system
Reagents | Usage (μL) |
2×ChamQ Universal SYBR Qpcr Master | 10 |
Primer F(10uM) | 0.5 |
Primer R(10uM) | 0.5 |
Template DNA/cDNA | 1 |
ddH2O | To 20 |
2.3.3 Mix the above system to make MIX, add it to the qPCR tube and mix until there are no bubbles in the tube.
2.3.4 Perform real time PCR in two steps. The reaction procedure is as follows:

2.3.5 Save the file data after the reaction is completed. Data analysis uses the double delta method (the calculation method is as follows), and the result expression is mean ± SD; use the one-way analysis of variance method to compare the differences between the two groups of data. P < 0.05 is a significant difference. ΔCt=Ct value of the target gene-Ct value of the internal reference gene; ΔΔCt=ΔCt of each sample-average ΔCt of the control group; 2-ΔΔCt reflects the relative expression level of the target gene of each sample relative to the control group
3. Result Example

The figure above shows the amplification curve of qPCR. The further back the curve is, the larger the Ct value is.
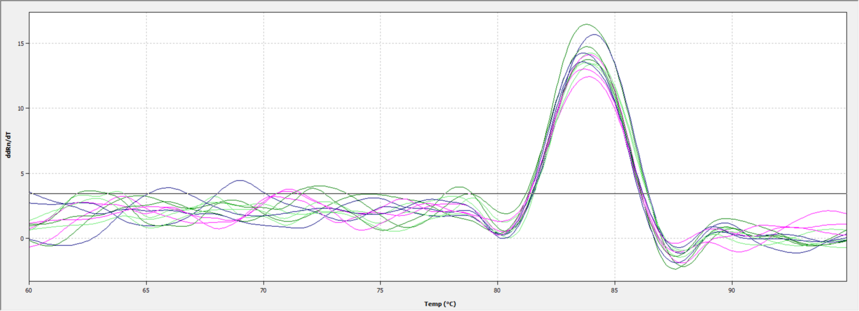
The figure above shows the melting curve of qPCR. The single peak of the curve indicates that the product has high specificity.

The figure above shows the qPCR result. The higher the value, the higher the relative expression of the gene.
References
[1] Rio DC, Ares M Jr, Hannon GJ, Nilsen TW. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc. 2010 Jun;2010(6):pdb.prot5439.
[2] Rio DC, Ares M Jr, Hannon GJ, Nilsen TW. Nondenaturing agarose gel electrophoresis of RNA. Cold Spring Harb Protoc. 2010 Jun;2010(6):pdb.prot5445.
[3] Green MR, Sambrook J. Quantification of RNA by Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR). Cold Spring Harb Protoc. 2018 Oct 1;2018(10).
[4] Arya M, Shergill IS, Williamson M, Gommersall L, Arya N, Patel HR. Basic principles of real-time quantitative PCR. Expert Rev Mol Diagn. 2005 Mar;5(2):209-19.
[5] Thornton B, Basu C. Real-time PCR (qPCR) primer design using free online software. Biochem Mol Biol Educ. 2011 Mar-Apr;39(2):145-54.
[6] Guénin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L. Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot. 2009;60(2):487-93.