RIP experiment (RNA Binding Protein Immunoprecipitation Assay) is a technique for studying the binding of RNA and protein in cells. It is a powerful tool for understanding the dynamic process of post-transcriptional regulatory networks. It can be used to study miRNA regulatory targets, the interaction between RNA and RBPs (RNA binding proteins), etc.
1. Experimental principle
RIP can be regarded as a similar application of the commonly used chromatin immunoprecipitation ChIP technology. It mainly uses antibodies or epitope markers against target proteins to capture endogenous RNA binding proteins in the nucleus or cytoplasm, and separates RNA binding proteins and their bound RNA together through immunoprecipitation. After the RNA in the complex is purified, the RNA bound to the complex can be verified by q-PCR or sequencing analysis.
2. Experimental flow chart

3. Experimental steps
3.1 Obtain cell lysate
3.1.1 Wash the cells in the culture dish or culture bottle twice with cold PBS.
3.1.2 After adding cold PBS, scrape the cells with a cell scraper and collect them in a centrifuge tube.
3.1.3 Centrifuge at 1500 rpm, 4 ℃ for 5 min, discard the supernatant, and collect the cells.
3.1.4 Resuspend the cells with an equal volume of RIP lysis buffer as the cells, blow evenly, and let stand on ice for 20 min.
3.1.5 Aliquot 200 μL of cell lysis buffer into each tube and store at -80 ℃.
3.2 Prepare magnetic beads-antibodies
3.2.1 Pipette 50 μL of the resuspended magnetic bead suspension into each centrifuge tube, add 500 μL RIP Wash Buffer to each tube, and vortex.
3.2.2 Place the centrifuge tube on a magnetic rack, remove the supernatant, and repeat the wash once.
3.2.3 Resuspend the magnetic beads with 100 μL RIP Wash Buffer, add appropriate amount of antibody, and incubate at room temperature for 30 min.
3.2.4 Place the centrifuge tube on the magnetic rack, discard the supernatant, add 500 μL RIP Wash Buffer, vortex and discard the supernatant, repeat once.
3.2.4 Place the centrifuge tube on the magnetic rack, add 500 μL RIP Wash Buffer, mix well and place on ice for later use.
3.3 RNA-binding protein immunoprecipitation
3.3.1 Place the centrifuge tube in 3.2.4 on the magnetic rack, remove the supernatant, and add 900 μL RIP Immunoprecipitation Buffer to each tube
3.3.2 Rapidly thaw the cell lysate prepared in 3.1.5, centrifuge, and pipette 100 μL supernatant into the magnetic bead-antibody complex to make the total volume 1 mL.
3.3.3 Incubate at 4 ℃ for 3 h or overnight.
3.3.4 Centrifuge briefly, place the centrifuge tube on a magnetic rack, and discard the supernatant.
3.3.5 Add 500 μL RIP Wash Buffer, vortex and place the centrifuge tube on a magnetic rack, discard the supernatant, and repeat the wash 3-5 times.
3.4 RNA purification
3.4.1 Resuspend the magnetic bead-antibody complex with 150 μL Proteinase K Buffer and incubate at 55 °C for 30 min.
3.4.2 After incubation, place the centrifuge tube on a magnetic rack and aspirate the supernatant into a new centrifuge tube.
3.4.3 Add 250 μL RIP Wash Buffer to each tube of supernatant, then add 400 μL phenol:chloroform:isoamyl alcohol, vortex and centrifuge at 14,000 rpm for 10 min at room temperature.
3.4.4 Carefully aspirate 350 μL of the upper aqueous phase and transfer it to another new centrifuge tube. Add 400 μL of chloroform to each tube, vortex for 15 s, and centrifuge at 14,000 rpm for 10 min at room temperature.
3.4.5 Carefully aspirate 300 μL of the upper aqueous phase and transfer it to another new centrifuge tube. Add 50 μL of Salt SolutionⅠ, 15 μL of Salt SolutionⅡ, 5 μL of Precipitate Enhancer, and 850 μL of anhydrous ethanol (RNase-free) to each tube, mix, keep at -80 ℃ for 1 h to overnight, centrifuge at 14,000 rpm, 4 ℃ for 30 min, and carefully remove the supernatant.
3.4.6 Rinse once with 80% ethanol, centrifuge at 14,000 rpm, 4 ℃ for 15 min, carefully remove the supernatant, air dry, dissolve with 10-20 μL of DEPC water, and store at -80 ℃. It can be used for subsequent q-PCR verification or sequencing analysis.
4. Result Example
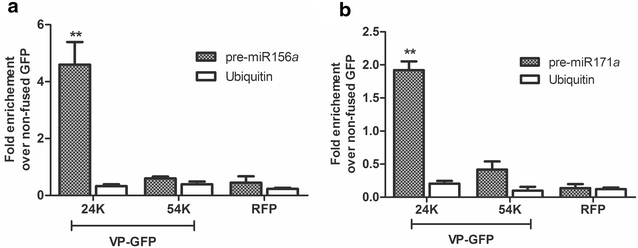
RIP analysis of precursors associated with 24K or 54K GFP fusion proteins of Citrus psoriasis virus (CPsV) in N. benthamiana. RT-qPCR was performed to determine the accumulation levels of pre-miR156a (a) or pre-miR171a (b) in N. benthamiana
References
[1]. Gagliardi M, Matarazzo MR. RIP: RNA Immunoprecipitation. Methods Mol Biol. 2016;1480:73-86.
[2]. Jayaseelan S, Doyle F, Tenenbaum SA. Profiling post-transcriptionally networked mRNA subsets using RIP-Chip and RIP-Seq. Methods. 2014 May 1;67(1):13-9.
[3]. Zambelli F, Pavesi G. RIP-Seq data analysis to determine RNA-protein associations. Methods Mol Biol. 2015;1269:293-303.
[4]. Nicholson CO, Friedersdorf MB, Bisogno LS, Keene JD. DO-RIP-seq to quantify RNA binding sites transcriptome-wide. Methods. 2017 Apr 15;118-119:16-23.