RNA pull-down is one of the main experimental methods for detecting the interaction between RNA binding proteins and their target RNA. This experiment can enrich and identify or verify proteins that interact with the target RNA.
1. Experimental principle
RNA pull-down is mainly used to detect the interaction between protein and RNA. RNA pull-down uses in vitro transcription to label biotin RNA probes, which are then incubated with cytoplasmic protein extracts to form RNA-protein complexes. The complex can bind to streptavidin-labeled magnetic beads and thus be separated from other components in the incubation solution. The proteins eluted from the magnetic beads are collected, and the proteins that interact with RNA can be determined and verified by mass spectrometry and Western Blot.
2. Experimental flow chart
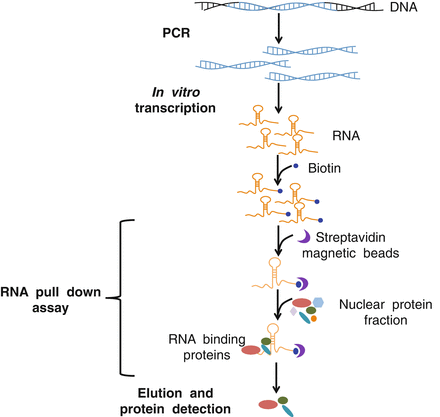
3. Experimental steps
3.1 Biotin probe labeling RNA
3.1.1 Take out the RNA 3' End Desthiobiotinylation kit, place the PEG and DMSO in the kit at room temperature, and melt the other components on ice;
3.1.2 Dissolve the RNA in DEPC water according to the content of the RNA mother tube, adjust the concentration to 10 μg/tube, and dissolve it with 10 μL DEPC water. At the same time, dissolve the control RNA in the kit as a negative control;
3.1.3 Take 5 μL of the control RNA and the target RNA, put them in a 200 μL PCR tube, add 1 μL DMSO to each tube, mix well, and place it on the PCR instrument at 85 ℃ for 5 min, and then immediately place it on ice for 5 min;
3.1.4 Prepare the reaction system according to the instructions of the kit
3.1.5 Mix the above reaction system, and then place it in the PCR instrument at 16 ℃ for 4 h to overnight. Prolonging the reaction time can improve the connection efficiency; after the connection reaction is completed, add 400 μL nuclease free-water to each tube;
3.1.6 Add 300 μL phenol chloroform to each tube to extract the successfully labeled RNA; after vortexing, centrifuge at the maximum speed for 15 min, and carefully transfer the upper aqueous phase to a new EP tube;
3.1.7 Add 10 μL 5 M NaCl, 2 μL glycogen, and 600 μL pre-cooled 100% ethanol, and place at -20 ℃ or -80 ℃ for precipitation. The precipitation can be overnight.
3.2 Prepare cell lysis buffer
3.2.1 Culture cells in a 15 cm culture dish. When the cells grow to 80-90%, discard the culture medium, wash with pre-cooled PBS three times, and aspirate the supernatant;
3.2.2 Add 200 μL cell lysis buffer to each culture dish;
3.2.3 Scrape the cells with a cell scraper, transfer to an EP tube, and place on ice for 30 min;
3.2.4 Centrifuge at 4 ℃, 3000 rpm, for 10 min;
3.2.5 Transfer the supernatant to a new EP tube and place on ice; add 200 U/mL RNase Inhibitor;
3.2.6 Take about 10-20 μL of supernatant to a new tube as experimental input.
3.3 RNA precipitation
3.3.1 Take out the RNA that has been precipitated overnight, centrifuge at 4 ℃ and maximum speed for 30 min. After centrifugation, the white precipitate at the bottom of the tube is RNA;
3.3.2 Carefully remove the supernatant, wash with 1 mL 70% ethanol, centrifuge at 8000 rpm for 10 min, then aspirate the liquid in the tube and dry the RNA in the air;
3.3.3 Add 20 μL nuclease free-water to each tube to dissolve the RNA, then place the RNA in a PCR instrument, denature it at 95 ℃ for 5 min, and then immediately place it on ice for use.
3.4 Magnetic bead pretreatment
3.4.1 Take 50 μL of magnetic beads into a centrifuge tube, place it on a magnetic rack, and aspirate the supernatant. Add 1 mL 20 mM Tris-HCl (pH7.5), wash the magnetic beads, place them on a magnetic rack, aspirate the supernatant, and repeat the wash once.
3.4.2 Wash 3 times with 800 μL 1×binding & washing buffer and remove the supernatant;
3.5 RNA binds to beads
3.5.1 Add 400 μL 2×binding & washing buffer to the beads;
3.5.2 Add 50 pmol labeled RNA to the previous step, and then add 380 μL DEPC water to make the final volume 800 μL;
3.5.3 Rotate slowly at room temperature for 30 min to allow the beads to fully bind to the RNA;
3.5.4 Place the tube in the previous step on the magnetic rack and remove the supernatant;
3.5.5 Wash 3 times with 800 μL 1×binding & washing buffer (containing RNase Inhibitor, 1 U/ μL) and remove the supernatant;
3.5.6 Wash the beads once with cell lysis buffer A and remove the supernatant. Be careful not to let the beads dry out.
3.6 RNA-protein interaction detection
3.6.1 Add 500 μL of cell lysis buffer to each tube of beads treated in the previous step; at the same time, add 1 U/μL RNase Inhibitor to each tube;
3.6.2 Rotate slowly at 4 ℃ for 2 h to allow the beads to fully combine with the cell lysis buffer;
3.6.3 Place on a magnetic rack to remove the supernatant, and wash the beads 5 times with 400 μL of freshly prepared cell lysis buffer A (+RNase Inhibitor+protease inhibitor);
3.6.4 Remove the supernatant, add 50 μL of Elution buffer, gently blow the magnetic beads evenly, incubate at 37 ℃ for 30 min, place on a magnetic rack, collect the supernatant, and perform Western blot detection or mass spectrometry detection.
4. Result example
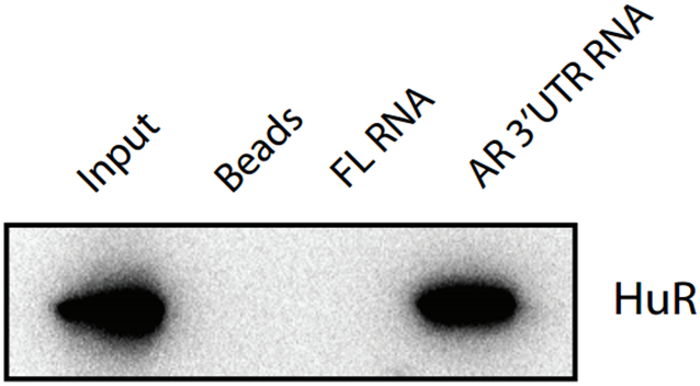
In this experiment, the beads group served as a negative control, and AR 3'UTR RNA was able to pull down HuR, indicating that there was an interaction between it and HuR.
References
[1]. Bierhoff H. Analysis of lncRNA-Protein Interactions by RNA-Protein Pull-Down Assays and RNA Immunoprecipitation (RIP). Methods Mol Biol. 2018;1686:241-250.
[2]. Popova VV, Kurshakova MM, Kopytova DV. [Methods to study the RNA-protein interactions]. Mol Biol (Mosk). 2015 May-Jun;49(3):472-81. Russian.
[3]. Barnes C, Kanhere A. Identification of RNA-Protein Interactions Through In Vitro RNA Pull-Down Assays. Methods Mol Biol. 2016;1480:99-113.