ChIP (Chromatin Immunoprecipitation, abbreviated as chromatin immunoprecipitation) is a technique for studying the interaction between proteins and DNA in vivo. It uses the specificity of antigen-antibody reaction to truly reflect the binding status of protein factors and genomic DNA in vivo. Chromatin immunoprecipitation technology generally includes cell fixation, chromatin fragmentation, chromatin immunoprecipitation, reversal of cross-linking reaction, DNA purification, and DNA identification.
1. Experimental principle
In the living cell state, the protein-DNA complex in the cell is cross-linked by fixing the cell, and it is randomly cut into small chromatin fragments within a certain length range by enzyme treatment. Then, this complex is precipitated by specific antigen-antibody reaction, and the DNA fragment bound by the target protein is specifically enriched. By purifying and detecting the target fragment, the information of protein-DNA interaction is obtained, and the genomic site where the protein binds is found.
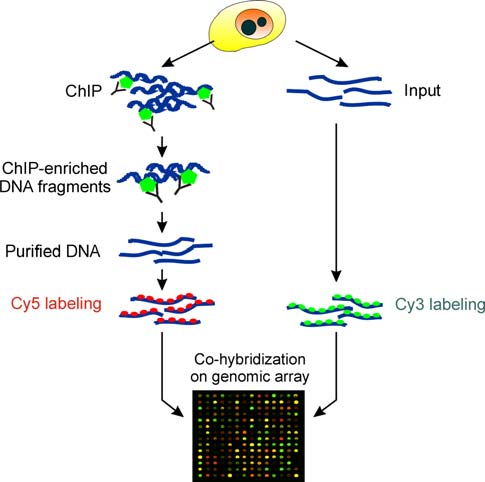
2. Experimental steps
2.1 Formaldehyde cross-linking and ultrasonic disruption of cells
Take out the cells cultured in the culture plate, add formaldehyde to make the final concentration of formaldehyde 1%; incubate at 37°C for 10 min, terminate the cross-linking, aspirate the culture medium, and wash the cells twice with pre-cooled PBS. Then use a cell scraper to collect the cells in two 15 ml centrifuge tubes, centrifuge and discard the supernatant, and perform ultrasonic disruption.
2.2 Removal of impurities and antibody incubation
After the ultrasonic disruption, centrifuge to remove insoluble substances, and add 900 μL ChIP Dilution Bufer, 20 μL × PIC and 60 μL ProteinA Agarose/Salmon Sperm DNA to 100 μL of ultrasonic disruption product. After inversion and mixing at 4 °C for 1 h, let it stand at 4 °C for 10 min to precipitate, centrifuge at 700 rpm for 1 min, and take the supernatant. Keep 20 μL of each as input. Add 1 μL of antibody to one tube and no antibody to the other tube. Invert at 4 °C overnight.
2.3 Test the effect of ultrasonic disruption
Take 100 μL of the product after ultrasonic disruption, add 4 μL 5 M NaCl, treat at 65 °C for 2 h to de-crosslink, and then separate half of it for phenol/chloroform extraction.
2.4 Precipitation and washing of immune complexes
After incubation overnight, add 60 μL ProteinA Agarose/Salmon Sperm DNA to each tube and invert at 4 °C for 2 h. After standing for 10 min, centrifuge and discard the supernatant, and wash the precipitated complex. Washing steps: add washing solution, invert at 4 °C for 10 min, stand at 4 °C for 10 min to precipitate, centrifuge at 700 rpm for 1 min, and discard the supernatant. After washing, start elution. De-crosslinking: Add 20 μL 5 M NaCl (final NaCl concentration is 0.2 M) to each tube, mix well, and de-crosslink at 65 °C overnight.
2.5 Recovery of DNA samples
After the cross-linking is resolved, add 1 μL RNAaseA (MBI) to each tube and incubate at 37 °C for 1 h. Add 10 μL 0.5 M EDTA, 20 μL 1 M Tris.HCl (pH 6.5), and 2 μL 10 mg/mL proteinase K to each tube. Treat at 45 °C for 2 h. Finally, recover the DNA fragments.
2.6 PCR, qPCR or sequencing analysis.
3. Example of results

In this result graph, the positive control group has clear bands without any other bands, and the negative control group is normal, indicating that the ChIP experiment results are reliable. The input control group has clear bands, indicating that the PCR amplification primers can be used to detect this indicator.
References
[1] Zhang, X.Guo,C. Chen, Y. Shulha, H.P. Schnetz, M. P.LaFramboise, T.Bartels, C. F.Markowitz, S. Weng Z.Scacheri, P. C. and Wang, Z. (2008) Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nat.Methods 5, 163–165.
[2] Haring M., et al. Chromatin immunoprecipitation:optimizationquantitative analysis and data normalization. Plant Methods. 11, 3-11 (2007).
[3] Nelson, J.D, Denisenko, O. & Bomsztyk, K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1, 179–185 (2006).