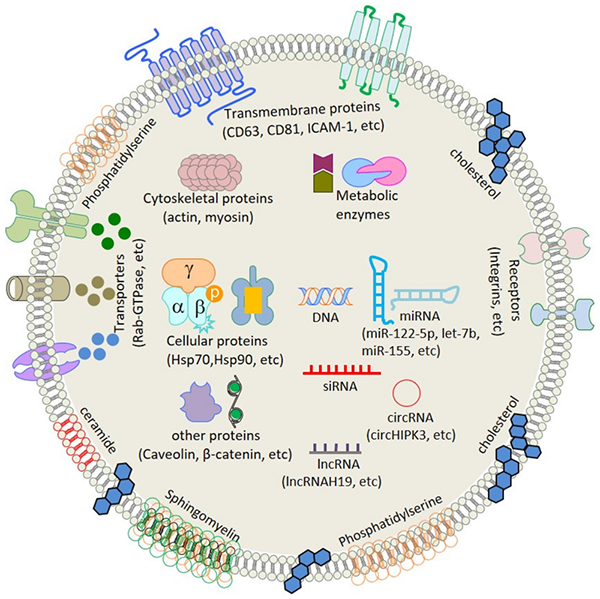
In exosome research, exosome identification is a key step after separation. Its purpose is to determine whether the sample contains exosomes, whether it can express specific exosome marker proteins, and whether it falls within the conventional particle size range of exosomes. In order to better conduct biological functional, morphological and biomarker research, it is necessary to identify the separated exosomes. Our identification methods mainly include transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA) and protein marker detection (western blotting).
1. Experimental principle
Exosome transmission electron microscopy can be used to identify the size and morphology of exosomes, and to observe whether there are exosome-like structures in the sample (usually a saucer-shaped or hemispherical shape with one side concave); the particle diameter analysis system can be used to measure and count the diameter distribution of particles in the sample to ensure the original state of the exosomes; after the exosome protein is lysed, the protein markers CD9, CD63, and TSG101 are detected to determine whether there is expression of related proteins in the exosomes.
2. Experimental steps
2.1 TEM identification
First, the exosomes are fixed with glutaraldehyde, and then 5-10 μL of exosome solution is added to the copper mesh, adsorbed at room temperature for about 10 minutes, and the excess liquid is carefully absorbed with filter paper. Then 10 μL of 2% phosphotungstic acid solution (pH=6.5) is added to the copper mesh, and the exosomes are stained at room temperature for 2 minutes. The excess staining liquid is carefully absorbed with filter paper, and the copper mesh is dried at room temperature and tested on the machine.
2.2 NTA identification
The sample volume of separated exosomes is generally between 2 μL and 100 μL. The zetaview software can analyze and obtain the particle concentration within a specific particle size range.
2.3 Protein marker detection
The exosomes were tested for three markers, CD9, CD63, and TSG101, according to the Western bolting experimental steps.
3. Result example
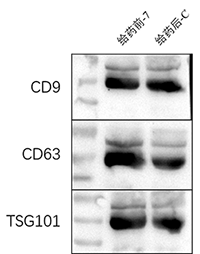
Western boltting test results showed that the extracted exosomes could successfully detect the expression of marker proteins CD9, CD63, and TSG101.
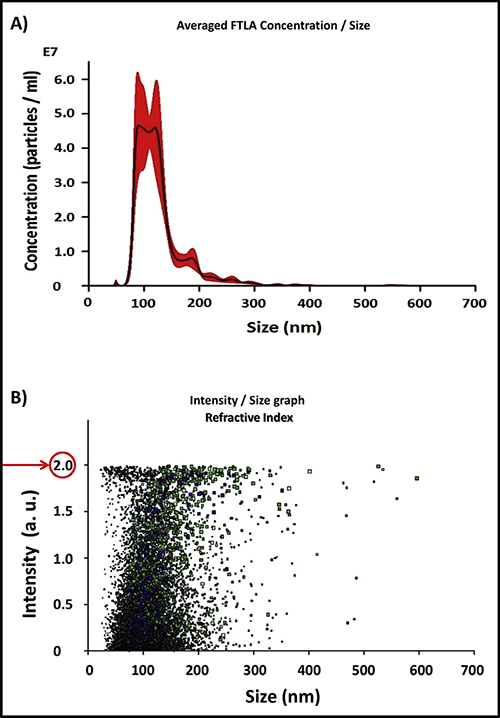
The NTA results above are exosomes released from human macrophages treated with AuNPs, showing a summary of exosome distribution, concentration (absolute counts), and refractive index.
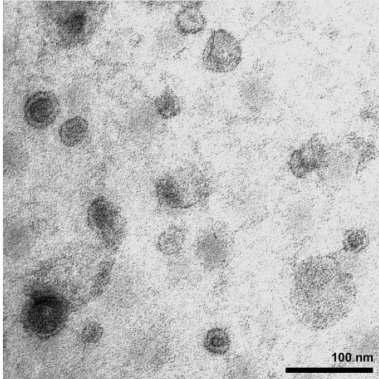
The extracted exosomes were examined by electron microscopy to determine their morphology and size and were identified as exosomes.
References
[1] Pluchino, S. & Smith, J. A. Explicating exosomes: reclassifying the rising stars of intercellular communication. Cell 177, 225–227 (2019).
[2] Mehdiani A, Maier A, Pinto A, Barth M, Akhyari P, Lichtenberg A. An innovative method for exosome quantification and size measurement. J. Vis. Exp. (95), 50974 (2015).
[3] J.L. Welton, S. Khanna, P.J. Giles, et al., Proteomics analysis of bladder cancer exosomes, Mol. Cell Proteomics 9 (6) (2010) 1324e1338.
[4] Hawker, G. A., White, D. & Skou, S. T. Non-pharmacological management of osteoarthritis. Osteoarthr. Cartil. 25, S4 (2017).